Enhancing Prenatal Screening with Low-Pass Genome Sequencing | BGI Insight
2023-10-18
Introduction:
The importance of accurate prenatal diagnosis in preventing birth defects cannot be overstated. Traditional karyotyping (a test to examine chromosomes in a sample of cells) dates back to the late 1960s and is well-established, but advancements in technology offer new options.
A new study led by Dr. Sun Yan, Dr. Lijie Song, Dr. Xueqin Guo at BGI Genomics R&D Division and published on Journal of Medical Genetics sought to validate Low-Pass Genome Sequencing (LP GS) as an alternative to Chromosomal Microarray Analysis (CMA) for prenatal diagnosis. Additionally, it explored the critical factor of sequencing depth in LP GS, which has been a gap in previous research.
Method and Findings:
The study analyzed 375 amniotic fluid samples and compared the diagnostic performance of LP GS to CMA. Surprisingly, both methods achieved the same diagnostic yield of 8.3%. In cases with negative CMA results, LP GS outperformed by revealing six additional Copy Number Variations (CNVs - Genomic alterations that result in an abnormal number of copies of one or more genes.) of uncertain significance.
Three key points to highlight:
1. This result emphasizes the robustness of LP GS in detecting genetic abnormalities, potentially missed by CMA.
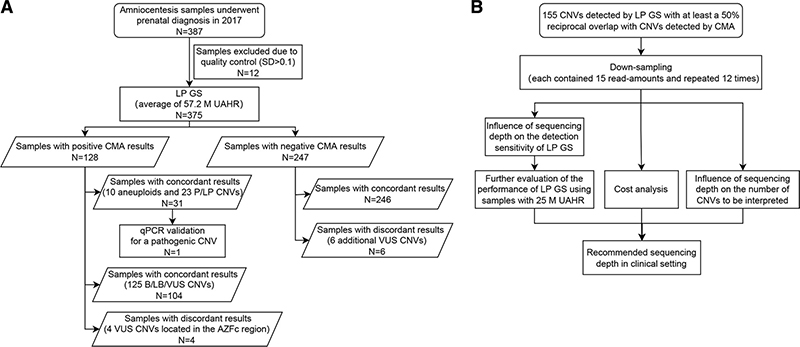
Figure 1 Flowchart of the study design. (A) Comparison of the performance of LP GS with that of CMA for diagnosing 375 amniotic fluid samples. (B) Depth evaluation of LP GS. CMA, chromosomal microarray analysis; CNV, copy number variation; LP GS, low-pass genome sequencing; P/LP, pathogenic/ likely pathogenic; UAHR, uniquely aligned high-quality read; VUS, variant of uncertain significance.
2. The study found out that the size of CNVs significantly impacted Low-Pass Genome Sequencing detection sensitivity. Small CNVs and those situated in specific regions, like the azoospermia factor c (AZFc) region of the Y chromosome, were more sensitive to variations in sequencing depth. In contrast, larger CNVs were less affected by sequencing depth.
3. The research recommended a sequencing depth of 25 million Uniquely Aligned High-Quality Reads (UAHRs) as optimal for detecting most aneuploidies and microdeletions/microduplications. This sequencing depth strikes a balance between detection sensitivity, cost-effectiveness, and interpretation workload.
These findings add crucial validation and data regarding LP GS as a first-tier prenatal diagnostic method in real clinical settings, highlighting its pivotal role of sequencing depth in LP GS and establishes the 25 million UAHRs threshold.
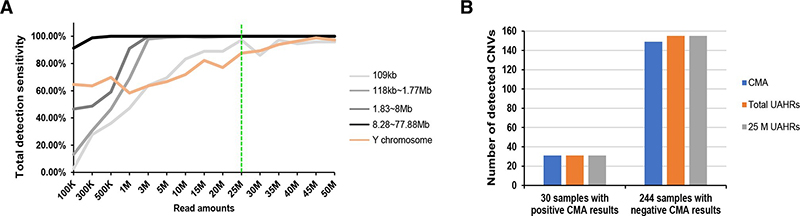
Figure 2 Depth evaluation of LP GS. (A) Total detection sensitivity of LP GS for 155 CNVs. The dotted green line shows the optimal UAHRs (25 M). (B)Evaluation of the performance of LP GS using samples with 25 M UAHRs. For the 30 samples with positive CMA results, the left bar figure shows the number of P/LP CNVs detected by CMA (blue bar), the number of concordant CNVs detected by LP GS using total UAHRs (orange bar) and samples with 25 M UAHRs (grey bar) compared with CMA. For the 244 samples with negative CMA results, the right bar figure shows the number of VUS CNVs (>100 kb) detected by CMA (blue bar), LP GS using total UAHRs (orange bar) and LP GS using samples with 25 M UAHRs (grey bar).
Enabling better decisions, reducing genetic abnormalities risks
Prenatal diagnosis is instrumental in preventing birth defects due to chromosomal abnormalities. This research findings reveal that LP GS can be a robust and cost-effective alternative to CMA in prenatal screening. It not only matches CMA's performance but offers a vital advantage in detecting additional CNVs. This enhanced capability can improve clinical decision-making and reduce risks associated with undetected genetic abnormalities, ultimately promoting safer pregnancies.
Enhancing Research and Clinical Outcomes:
These findings provide a significant leap forward in prenatal care. The ability to accurately detect aneuploidies and microdeletions/microduplications enable reducing birth defects, more accurate patient care and better clinical decision-making
About BGI Genomics:
BGI Genomics, headquartered in Shenzhen, China, is the world's leading integrated solutions provider of precision medicine. Our services cover more than 100 countries and regions, involving more than 2,300 medical institutions. In July 2017, as a subsidiary of BGI Group, BGI Genomics (300676.SZ) was officially listed on the Shenzhen Stock Exchange.
Read more:
Genomic Sequencing as a First-Tier Screening Test for Newborns
How cfRNA Could Revolutionize Preeclampsia Diagnosis
Whole-genome sequencing is a more comprehensive prenatal test
How can whole genome sequencing and carrier test help screening for birth defects
What 10 years of experience in non-invasive prenatal testing applications can tell us